Experimental evolution of cooperation and
conflict: A model
system
For my dissertation project I use a model system with viruses to experimentally study the evolution of cooperation and conflict. I developed this system with my advisor, Jim Bull. The experiments described below use non-lethal viruses to directly simulate the processes of cooperation and conflict between two individuals. I co-infect two bacteriophage species in the same line of bacterial cells and expose them to different selective regimes. In the main experiment I enforce the coexistence of the two phage during certain parts of their life histories, but allow independent reproduction. Since evolution occurs on the order of days or weeks in viral populations, I can test hypotheses of between-species cooperation in a rather timely manner. Adaptive outcomes in these in vitro evolution experiments are analyzed at the phenotypic and the genotypic level.
Background and application to nature
Cooperation is a challenging problem to evolutionary biologists because
it is at odds with the “selfish gene view,” which holds that
genes (or more generally any unit capable of evolution) evolve selfishly
towards increased propagation. Cooperation is a central theme in biological
evolution at many taxonomic and organizational levels. The cooperation
between early replicators was likely a critical step in the origin of
life, was necessary for the endosymbiotic origins of eukaryotes, as well
as the diverse symbioses we observe in nature.
Yet, with cooperation there is always conflict. Conflict is likely to be present between any entities that can reproduce independently, including many interactions generally considered cooperative. Examples range from conflict within social groups, parent-offspring conflict, sexual and sibling conflict, to inter-cellular and intragenomic conflict. The interplay of cooperation and conflict becomes of particular interest when considering evolutionary “integration” events (Maynard-Smith and Szathmary 1995). Integration occurs where entities that could once reproduce independently become joined into one replicating unit. Examples include the putative association of replicators into proto-cells, genes into chromosomes, protists into multicellular organisms, and integration of different prokaryotes into the first eukaryotes. Similarly, many endosymbiotic organisms have free living stages that alternate with intimate co-existense with other symbiont genotypes within their common host. The challenge is to understand how cooperation can evolve and be stable between two entities that are each selected to maximally self propagate. However without experimentation, we are limited to studying past evolutionary transitions. This work will directly address this paradox (the evolutionary origin of cooperation) through experimental evolution.
Synopsis of Experiment
Infection and vertical transmission: Two non-lethal bacteriophage
species, f1 and IKe, are co-infected into a special cell-line of Eschershia
coli. Each phage has been engineered to contain a different antibiotic
resistance gene, allowing selection for the presence of either (or both)
of the phage. After the initial co-infection, both antibiotics are added
so only co-infected cells survive and reproduce. Infection of two phages
into one cell potentially leads to conflict; each of these phages uses
similar resources within the cell and their gene products may be similar
enough to interfere with each other's reproduction.
Once antibiotics are present, however, phage may only contribute to the next generation via vertical transmission within the co-infected cell lineage, since no new infections can occur. The co-infected culture is allowed to grow to saturation. During this long phase of vertical transmission (of both phages) there is selection for cooperation between the two phage. Each can only reproduce in the presence of the other, and evolution of cooperation between them can maximize their reproduction.
Phage production: Once the culture reaches saturation, coinfected cells are isolated from the culture with centrifugation and washing, then are allowed to grow again for one hour in the presence of both antibiotics. This allows the production of phage progeny for the next passage. Although the co-infected cells have been producing phage all along, this process isolates the phage produced by cells that survived the overnight growth phase. The cells in this culture are then heat killed, which leaves the phage unharmed. These phage are then used to infect naive cells of the identical genotype (as above) and the process is iterated. Fifty iteractions (passages) of the above experiment have been completed.
Genotypic and Phenotypic analysis
The genotypic aspect is rather straightforward. I have sequenced both
of the bacteriophages before and after the evolution. More sequencing
and genetic manupulation will elucidate the order and fitness effects
of the mutations that occured.
Phenotypic evolution is more complex, though several aspects of the phage interactions can be measured over the evolutionary experiment. Cooperation and conflict between the phage can be measured in at least two simple ways.
1. Per-cell output of each phage: A measure of conflict. By estimating each phage's output per-cell during the phage production phase I can track their output ratios over time. Output ratio indicates whether one phage attains high production at the expense of the other, or if both can raise production cooperatively.
2. Fitness of co-infected cells: A measure of cooperation. During the overnight phase of vertical transmission of both phages within the cell line each phage must cooperate with the other to maximize cell division. Evolution of this joint trait can be meausred by tracking fitness of co-infected cells over the experiment.
Initial Results
Genotypic analysis is well under way, and phenotypic analysis has just
begun. The initial data indicate that major evolutionary changes have
occurred in both phages in the course of the experiment. Most dramatically,
one of the phage (IKe) evolved a major reduction in genome size. The new
"IKe particle" phage is completely reliant on the other phage
(f1) for its own reproduction and packaging (because it has lost critical
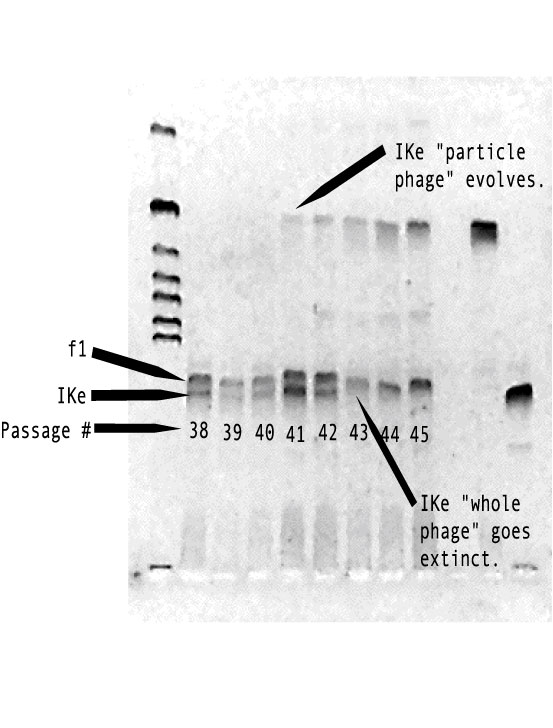
genes for these functions).The below figure shows the the evolution of
the "IKe particle" and that it spreads throughout the culture
driving the whole IKe extinct. Each lane in this gel shows the bacteriophage
DNA extracted from the co-infected culture of cells from passages 38-45.
This is akin to the evolutionary origin of chromosomes, where genes that
could once replicate independently become joined and can only replicate
as an integrated unit.
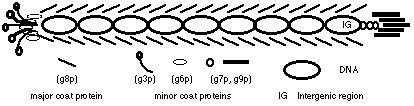
The Bacteriophage players and genetic details:
The model system employs the two filamentous coliphages f1 and IKe. Filamentous phages are composed of single stranded DNA encased in a thin protein filament. They establish permanent non-lethal infections on piliated, gram-negative bacteria only, and reproduce by extrusion through the bacterial cell wall without lysis (Model & Russel 1988). The length of the protein filament is determined by genome size of the phage, allowing relatively easy genetic manipulations including insertion of non-phage DNA (Messenger et. al. 1999). IKe and f1, although sharing only 55% sequence identity (Peeters et. al. 1985), have the same 10 genes in identical arrangement and share other characteristics suggestive of common ancestry (Model & Russel 1988). IKe and f1 infect different types of piliated Eschereshia coli cells; f1 requires hosts with an F-episome (expressing the F pilus) while IKe requires the N pilus expressed by an IncN episome (Bradley 1979). IKe and f1 interfere with each other within the cell, and have a 100-fold fitness loss when coinfecting cells (Russel 1992).
Each phage was engineered in our lab to include resistance to the antibiotics kanamycin (Kn) on IKe and chloramphenicol (Cm) on f1. In this way I can impose selection so that only cells coinfected with both IKe and f1 are able to grow, and thus contribute phage to the next passage. These gene inserts are also useful for assaying the presence and frequencies of each phage in culture by plating with either or both antibiotics present. Each phage can also be isolated by employing different cell lines that express just the F pilus or N pilus, and can only be infected by f1 or IKe respectively. Genomes of IKe and f1 are both under 7,000 nucleotides in length and have been sequenced in our lab. Their gene functions are relatively well understood, so phage adaptation is readily studied at the genotype level. A final benefit of this system is that genomes can be stored indefinitely throughout the experiment by freezing, and results can be replicated to observe if the selection leads to one or many possible outcomes.
.jpg)
.jpg)
.jpg)
.jpg)

References: -Bradley, D. E., 1979. Morphology of pili determined
by the N-incompatibility group plasmid N3 and interaction with bacteriophage
pr4 and bacteriophage IKe. Plasmid. 2, 632-636.
|